magazine_ Interview
Meeting C. elegans
How a tiny, wriggling worm is enabling discoveries into the mechanisms of Parkinson’s Disease.
The nematode worm Caenorhabditis elegans has emerged as an important animal model in various biomedical studies. Here we look at the role it can play in understanding Parkinson’s disease.
Transparent worms, also known as nematodes, are everywhere. These are mostly microscopic roundworms that have adapted to a spectacular range of environments, from Antarctic soil to the inside of your gut. They are present in so many microenvironments that Nematologist Nathan Cobb, once predicted “if all the matter in the universe except the nematodes were swept away, our world would still be dimly recognizable.” Recently, Nematode worms have been in the news, from the discovery of a 46,000-year-old nematode locked in the Siberian permafrost brought back to life, to the carnivorous oyster mushroom that feeds on nematodes by injecting them with a nerve gas, to the fact that as one of the most prolific species on our planet it should come as no surprise that the Florida manatee’s skin hosts about half a million of them.
C. elegans is just one of the species, but as you will read, a very interesting and important one. Measuring just one millimeter from head to tail, C. elegans usually lives in the soil and feeds on bacteria and other micro-organisms. Most adults are hermaphrodites, with both eggs and sperm, and produce several hundred offspring through self-fertilization. In fact, one nematode can produce 1000 progeny. In a week.
Roman Vozdek is a Biochemist, biomedical engineer, and the Eurac Research Institute of Biomedicine’s first and only nematologist. After having set up Bolzano’s first C. elegans lab we talk to him about how these microscopic marvels can help understand the mechanisms around Parkinson’s Disease.
What's a typical day for you?
Roman Vozdek: First thing I usually do when I get to the lab is checking the plates that I'm interested in. If I crossed worms two days before, I check if they crossed or not. Sometimes they don't. So if they don't, I have to set up the mating again.
With candles and romantic music?
Roman: Ha. They actually listen to Pearl jam a lot. Anyway, for the cross all I need to have are males from one strain. I put them on the plate with the hermaphrodite of another strain and then check to see if there is progeny of the two original genotypes. I usually check the genotype of these worms by doing a PCR, or sometimes this can be done by microscopy relatively fast. Next, I look at other plates. If I’m doing some experiments, I check how the experiment is going, investigate whether the phenotype that I expected is being exhibited. I also use some biochemical techniques to investigate gene functions and basically spend all day in the lab doing what needs to be done.
How do you record your findings?
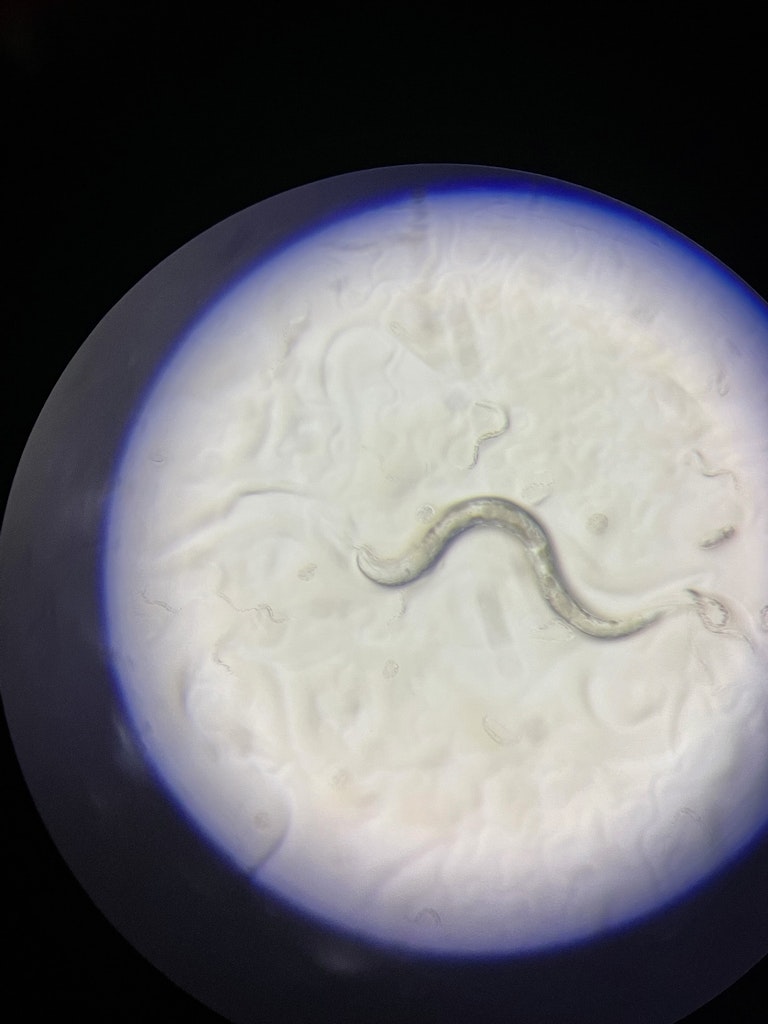
Roman: I take photos of the worm or video of the worm’s behavior. Worms exhibit different behavior under different conditions. You can study how fast they crawl and how they respond to stress. Stress factors can be manipulated chemically, mechanically or with temperature. You can see whether the worms have the foraging phenotype or if they lack the sense for finding the food. You can analyze how they swim, and respond to liquid. Although each worm is very small, they are quite thick for detailed imaging. And sometimes when you photograph the whole organism, you can't see the details of each tissue. Because the worm is a complete organism. Muscles, neurons, hypodermis, etc. But you can take a detailed image with something called confocal microscopy when you take tissue scans and then build a three-dimensional image. This is all done when the worm is immobilized, because otherwise they crawl all over the place. The important thing is they are alive. So, using C. elegans we can study the biology in-vivo, which is the study of something in a living organism.
C. elegans: in-vivo
Video: Eurac Research | Annelie Bortolotti
How do you get hold of these? Where do these worms actually come from?
Roman: They live in most soil types and are found on every continent. You just need to look under a microscope to see them. The most famous C. elegans was isolated in Bristol in the UK, the original wild strain of C. elegan*s, that is being used as a reference strain in all research labs studying *C. elegans. But now we know there are tons of wild strains because you can isolate these worms anywhere in the world. A worm community established a C. elegans depository that collected the worms generated in the different labs around the world and that distribute them if there’s interest. You can also share the strains directly with other labs. The worm community is just very generous and friendly. And so, you get the worms from these labs or a depository, all you need to do is be working in a registered C. elegans lab. When I started to work with worms, it was $7 for one strain, now it’s around be $10. They send you Petri plate with hundreds of worms on it because they need to survive shipping – they’re on the way weeks usually because they get stuck at customs. But for the worms, it's not a problem as they can live months without food. One strain means they're all identical having a defined genotype –the DNA is the same in each individual. I have already ordered more than 100 different strains and made more than 300 myself over the past four years.
How do you get the Parkinson's gene into the worm?
Roman: In our case I used human gene encoding protein called alpha-Synuclein, which when aggregated becomes toxic for neurons, and which is also associated with familial forms of Parkinson’s. I inserted this human gene between the pieces of C. elegans DNA that make sure the gene will be active only in specific C. elegans neurons. We can inject DNA into the gonad of the worm, whose eggs will take it as their own. The worms carrying this human gene do suffer from neurodegeneration but won’t have classic Parkinson's symptoms, such as tremors, because that is related with human biology.
And why in worms exactly?
Roman: It started in Prague when my PhD study focused on the role of few C. elegans genes in hydrogen sulfide metabolism. We discovered really interesting and unexpected functions of these genes and I just realized how cool the worm is for research and wanted to continue to study more of C. elegans biology. When I finished my PhD, I moved to the States where I worked on a study with actual worms, not just their DNA or their proteins. We were working on a protein which remarkably rescues, or rather promotes survival of cultured dopaminergic neurons., Dopaminergic neurons are the ones that die when you have Parkinson’s disease. We were using C. elegans and working on how this protein, the mechanism, really rescues these neurons and I started to be interested in neurodegeneration. Naturally I contacted Andrew Hicks, Vice Head of the Institute for Biomedicine, who was working with the cells from actual patients, to find out if he would be interested in using worms to understand more of the pathology. An advantage of C. elegans is that it is a complex living model to study the basic biology of Parkinson’s disease-associated genes. You find out what these genes do in a way that is simple, easy and inexpensive. Then you can look at the human biology. Is it the same or not? Much of the biology present in worms is also present in humans.
I heard there was a worm that was brought back to life after 46,000 years in permafrost, a nematode.
Roman: So that's another big advantage. You can actually freeze them, as we do with bacteria regularly, different strains of bacteria. But with worms you can do the same, it's really big benefit of this model. You can freeze them and take them after several years and they’re still alive.
Can you explain the science and the end goal of this specific project?
Roman: The pathological hallmark of the Parkinson’s disease are the Lewy bodies, abnormal build ups of protein that develop in the brain and these Lewy bodies contain aggregated protein called alpha-Synuclein. As I mentioned before, when the gene for alpha-Synuclein is in the worm and you activate the gene in the neurons, they degenerate. So, we thought by using a worm model of Parkinson's disease, with degenerated dopamine-synthesizing neurons, we could study the role of different genes or the effect of different metabolites and chemicals on neurodegeneration. This way it could be possible to discover fundamental neuroprotective mechanisms that are evolutionarily conserved in humans as well. However, there was no strain in any of the depositories or labs in which you could monitor how this protein aggregated or where it was expressed within the neuron. So, I first made this worm that had a fluorescent tag on the alpha-Synuclein. I use colors, green, to visualize the Synuclein, so you can see what it does, where it aggregates in the neuron, where it's expressed, under what conditions it doesn't aggregate, etc. And red, to view actual neurons to show exactly where the neuron is and what its morphology is. This strain has become quite popular and is being used in several labs around the world. Like this, you can visualize organelles, cells, entire tissues. You can visualize the proteins. You can go really into detail with these fluorescent tags in a living organism. By the way, the Fluorescent Green Protein is actually a gene from jellyfish. In our laboratory, I use several C. elegans models for Parkinson’s, like the one I mentioned, and study how different genes associated with the disease, play a role in these worm models. We have identified some genes that are neuroprotective or neurotoxic and so with the colleagues at the Institute we will have to follow up and evaluate the role of these genes in human cellular models to see which of these genes will have a potential to be therapeutic targets for Parkinson’s Disease.
Nematodes and Nobel Prizes
In 2002 John Sulston, Sydney Brenner, and Bob Horvitz, shared the Nobel Prize in Physiology or Medicine “for their discoveries concerning genetic regulation of organ development and programmed cell death” or how a many-celled body trades-off between cell division and cell suicide. Studies of the nematode worm, Caenorhabditis elegans, led to a fundamental reappraisal of the importance of genes involved in the deliberate killing of certain cells during normal development. To study these mechanisms, Sydney Brenner, investigated the transparent nematode worm to understand how genes control development from a fertilized egg. In a series of scientific publications, the team mapped the lineage of every nematode cell and described the visible steps in programmed cell death, demonstrating the first genetic mutations involved in the process and identifying the first two bona fide “death genes” - ced-3 and ced-4 which the human genome also contains.
Some organisms, like jellyfish ‘glow’ because they emit a shimmering light that has been called Green Fluorescent Protein (GFP). In 2008 Martin Chalfie also garnered the lauded prize for discoveries that enabled him to insert the GFP gene into the C. elegans and in doing so was able to ‘color’ six individual cells that could then be tracked.
Why C. elegans?
Caenorhabditis elegans is the perfect subject for a study of how a complete organism can be made from the genetic code of DNA. Each mature worm has exactly 959 cells and due to a rapid development from egg to adult which can be easily viewed through a microscope, a full life cycle can be studied by mapping what happens to each nematode cell during development. This nematode can grow from egg to adult in just three days. Scientists like the nematode because it can be easily kept in the laboratory, feeds very happily on bacteria growing in a petri dish – and has a remarkably simple body plan. This simplicity helps understand how a multi-celled organism is created from genetic information encoded in a DNA molecule.
About the Interviewed
Biochemist and biomedical engineer Roman Vozdek gained experience in laboratories for C. elegans nematodes research at MIT and UCSF. He uses these roundworms finds novel therapeutic targets for the treatment of the diseases associated with oxidative stress, including Parkinson’s Disease. After obtaining a Masters in Biochemistry and Biomedicine, his Ph.D. and postdoctoral work identified genetic pathways and molecular mechanisms promoting cellular adaptation to hypoxia and oxidative stress in the roundworm C. elegans. He has been successfully funded by many grants and fellowships and has published in Nature communication, Science signaling, Frontiers in Aging Neuroscience. He established the C. elegans laboratory at Eurac research to leverage C. elegans genetics as a powerful discovery tool to identify novel mechanisms of neurodegeneration. His STANIMON project was funded by the international mobility grant of the Autonomous Province of Bolzano - CUP D55F21004130003.